Research
Overview
Most of the organisms we are familiar with (including ourselves) are diploids, which are called that because they (we) have two homologous copies of every chromosome. But some organisms, so-called “polyploids,” have more, sometimes many more. Species with four or six or even eight copies of each chromosome are quite common, but some species have many more – the record holder is currently the fern Ophioglossum reticulatum, a 96-ploid with 1440 chromosomes!
Polyploidy has played a major role in the evolution of genome complexity in all eukaryotic kingdoms but is especially prevalent among plants – many of our crop species (cotton, wheat, potato, blueberry, coffee, strawberry, to name a few), but also many wild ones, are polyploids. Polyploidy can play roles in habitat adaptation, invasiveness, and speciation. Newly generated polyploids could also be useful in crop improvement, as new polyploids often have strong resistance to abiotic stresses such as salinity and drought.
For understanding polyploid evolution, or for utilizing new polyploids in crop improvement, we run squarely into the issue that neo-polyploids have very low fertility. While not the only challenge neo-polyploids face upon genome duplication, it is likely one of the most pressing things a newly polyploid lineage must solve to survive. Our lab therefore focuses mostly on studying the evolution of fertility in polyploid lineages, though we are also interested in other challenges they may face to do with their cell size increase, and the evolved responses that solve neo-polyploid problems.
Our group uses varied approaches, including genetics, molecular biology, protein biochemistry and cytology to understand the molecular and mechanistic basis of how adaptive evolution can re-establish fertility and core cellular functions after genome duplication. We study genes we previously identified as having been under selection in a natural tetraploid lineage of our model, Arabidopsis arenosa. Knowing what was targeted by selection can help us understand both what challenges neo-polyploids face, as well as what solutions evolved in response to them (and in turn, the fact that a solution had to evolve, also allows us to interpret our lists of genes to come up with new hypotheses about what things constitute challenges in the first place).
Currently we are focused on three major areas:
Meiosis: Polyploids face the unusual challenge of segregating more than the usual pairs of homologs. Neo-polyploids have unstable meiosis, associated with low fertility, while evolved polyploids have (mostly) stable meiosis. We are studying the genes and mechanisms that got them there.
Pollen Tubes: We recently discovered that tip-growth of pollen tubes represents an additional major challenge to polyploid fertility. We are now working to understand why, and how the evolved polyploids “fixed” the issue. This likely involved both chromatin-regulation of vegetative nucleus activation as well as mechanical adjustment of tube growth. We are exploring both angles.
Chromatin: We seek to understand how chromatin architecture and regulation is affected by genome duplication, considering the three-dimensionality of the genome, and how arrangement of DNA within nuclei impacts gene regulation and how it relates to cell size. This may provide novel insights into the “nucleotypic effect” which has been recognized as a polyploid challenge that arising from scaling changes that arise from the cell size increase that accompanies genome duplication.
Research

What polyploids manage is remarkable! We know from decades of research that duplication of an entire genome can pose challenges to core processes like gene regulation, intracellular trafficking, ion homeostasis, and chromosome segregation, yet these organisms generally survive (except in mammals, it seems). Nevertheless, newly formed polyploids do face multiple challenges to their development, and especially, their fertility. Understanding how polyploid lineages that manage to successfully establish and evolve solutions to these initial challenges is both fundamentally interesting, while also possibly allowing wider use of polyploidy as a crop improvement tool. This is of interest because polyploids are noted not only for their adaptability, but also their strong resistance to a wide range of agriculturally relevant environmental stresses, such as salinity and drought.
The lab uses diverse approaches, ranging from genomics to protein biochemistry and cytogenetics (Figure 1), to gain a detailed mechanistic understanding of how polyploid lineages adapt to genome duplication. We mostly focus on genes identified in our prior work as having been candidate targets of selection during the evolution of the established tetraploid lineage of Arabidopsis arenosa (Hollister et al. 2012; Yant et al. 2013; Bohutínská et al. 2021).

A main focus of our research is thus to understand what it takes to be polyploid. In order to study how polyploid lineages might adapt to the fact that they are polyploid, our group has been, since the lab was originally founded at Harvard University in 2009, developing Arabidopsis arenosa (Figure 2) as a model system. This system has the strength that it occurs in nature both as an extant diploid, as well as a tetraploid lineage that has spread through much of Europe. We know the tetraploid lineage arose just once, in the Carpathian mountains, probably around 30,000 years ago or a bit more. In addition, we can generate new polyploids in the lab. This “three part” system allows us to study the callenges that arise with the genome duplication event (by comparing diploids with neo-tetraploids generated from them), as well as the solutions that the natural tetraploid lineage evolved in response (by comparing neo- and established tetraploids). In addition, our prior work on what genes were targets of directional natural selection in the tetraploid lineage help us formulate new hypotheses not only about what genes evolved novel alleles to alter biological processes in response to challenges, but also allows us to generate new hypotheses about what might be challenges that we had not previously thought of (we recently used this “reverse” approach to identify pollen tube tip-growth as an important challenge to polyploid fertility, which now constitutes a major research focus of the lab).
The main research directions of the lab are all framed by the overarching question: How do polyploids adapt to being polyploid?
Meiosis
It is well known that new polyploids face challenges in chromosome segregation during meiosis. This arises from the fact that each chromosome suddenly has more than the usual two copies of each homologous chromosome that can recombine and need to separate from each other in meiosis. This leads to challenges and can result in fertility reduction. A major avenue of research in the lab focuses on trying to understand what problems arise with chromosome pairing, recombination, and segregation in meiosis in neo-polyploids, and what novel alleles evolved to fix the problem. Ultimately, we hope to gain new insights into basic meiotic processes from understanding how they can be modified by evolution to answer the problems that genome duplication presents them with.
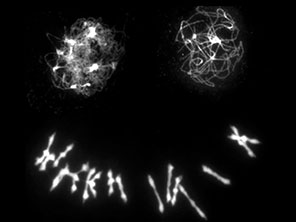
In our previous work, in collaboration with Nancy Kleckner (Harvard University) and Chris Franklin (University of Birmingham), we showed that neo-polyploid A. arenosa faces important challenges in chromosome synapsis and recombination. Problems are evident when we compare chromosomes in metaphase I of meiosis in newly generated A. arenosa tetraploids that we can generate using colchicine (a microtubule disrupting chemical) from the diploid, and compare this with the established tetraploid, which has generally stable meiosis (Figure 3).
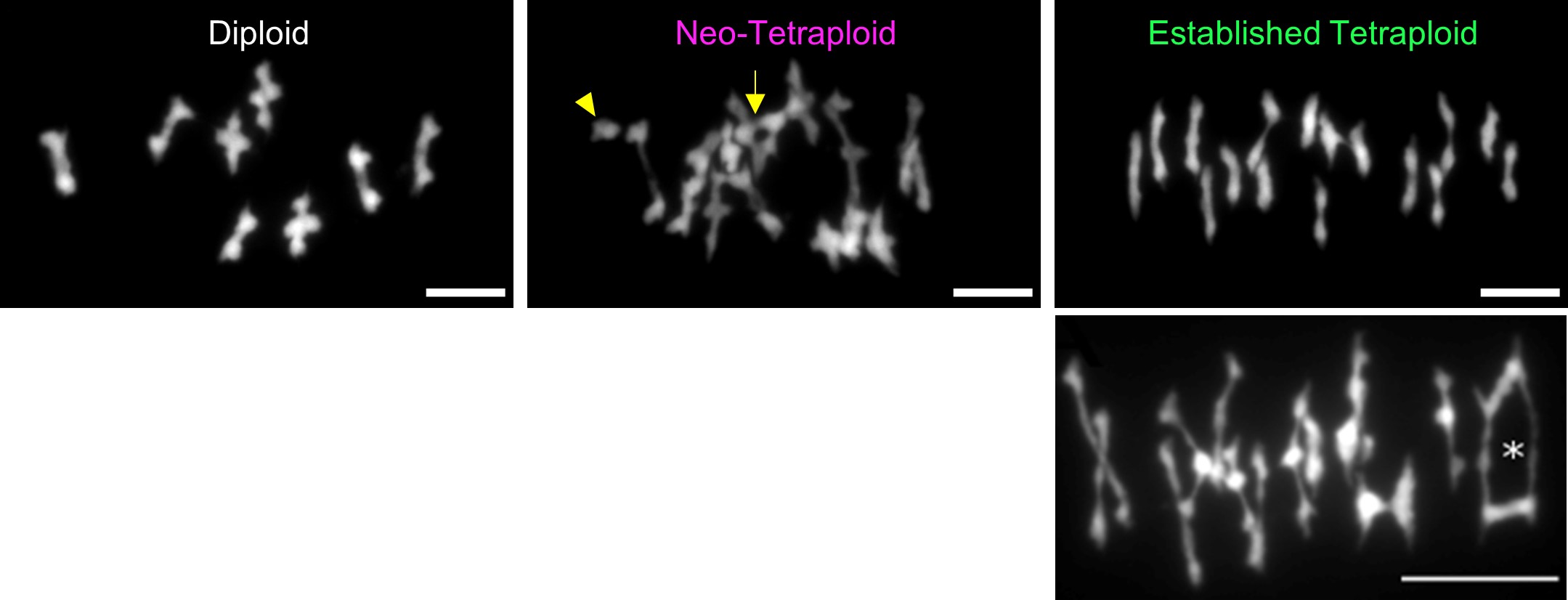
The neotetraploid A. arenosa, as has been reported in other species, shows extensive problems in meiosis including having unpaired chromosomes that will likely mis-segregate or be eliminated, and multivalents, which result from recombination events linking more than two copies of each homolog, can also cause problems for chromosome segregation. Multivalents are widely reported in the literature to be linked to low fertility in neotetraploids, and evolved tetraploids form far fewer than neotetraploids. We found evidence that a critical part of the solution that evolved in the established polyploids, is stronger crossover interference, meaning not only that crossovers are placed farther apart, but that they are also placed in arrangements on four-chromosome groups that are less likely to yield multivalents or univalents (See Morgan et al. 2021). Placement relative to synaptic partner switches (Figure 4) is important. Now we are attempting to understand how exactly the polyploid lineage achieved this elevated crossover interference and are working on candidate genes (from our previous scans for selection in polyploids) that affect processes like pairing and synapsis.
We hypothesized some time ago (Bomblies et al 2016) that reduced crossover rates could cause there to be fewer multivalents, as it reduces the likelihood of connecting multiple chromosomes. A recent study from the lab (Gonzalo et al 2023) showed that this prediction indeed plays out: reducing crossover rates in mutants in A. thaliana indeed reduced the frequency of multivalents in neo-tetraploid lines created from them.
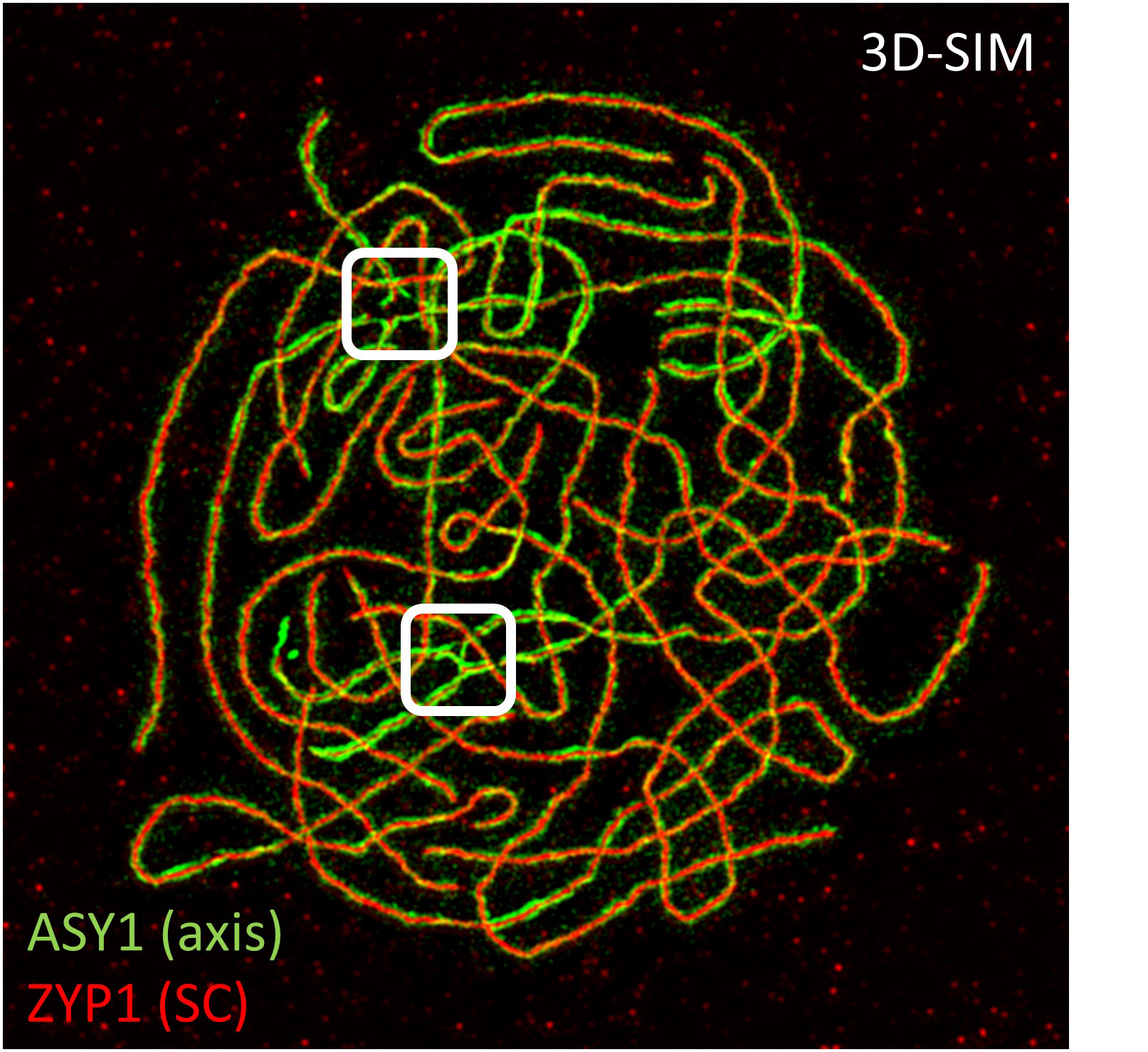
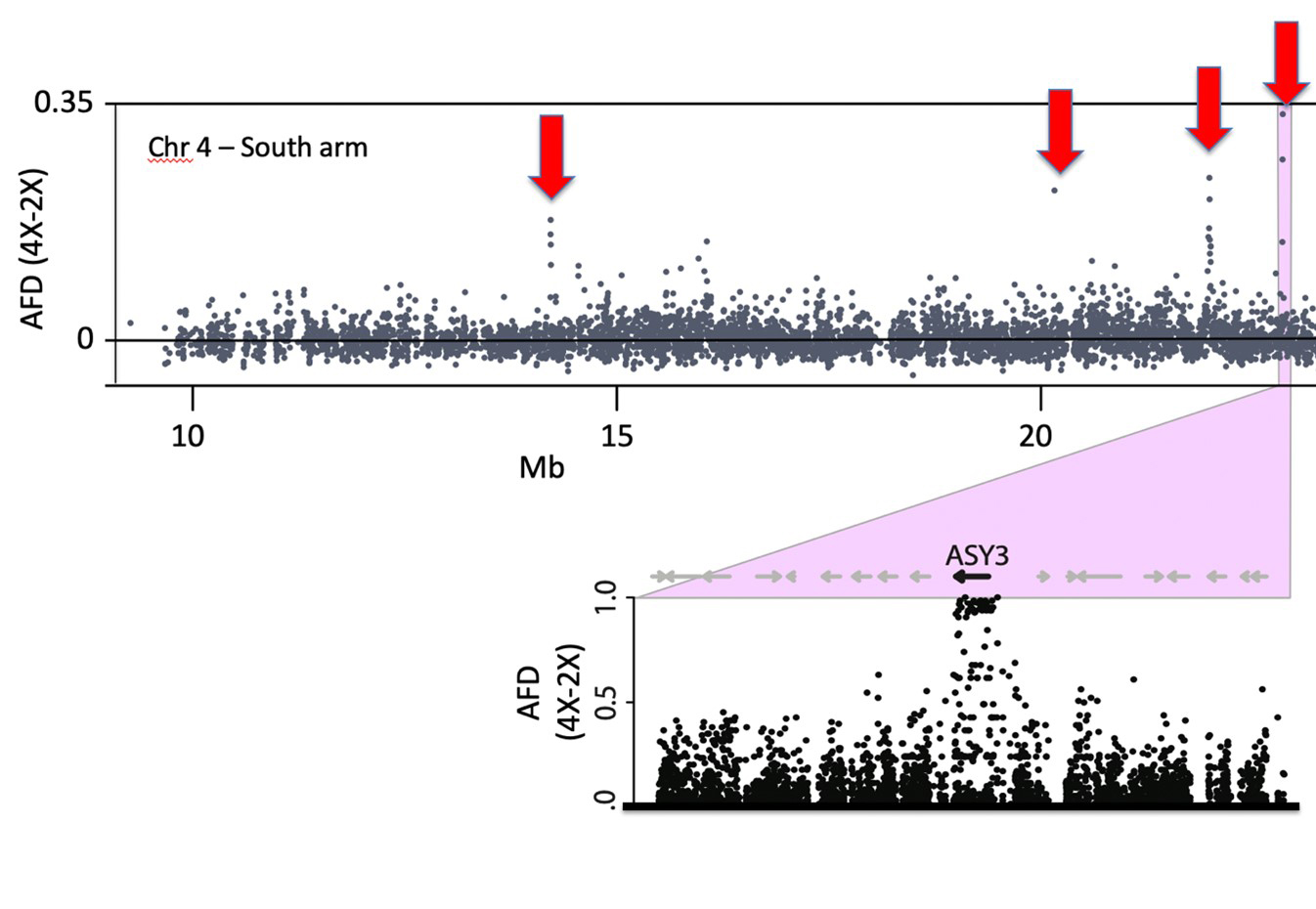
What are the relevant genes that drive meiotic adaptation in the tetraploid? In a set of genome scans for selection in the tetraploid lineage, we identified genes with clear evidence of having been targets of natural selection in the tetraploid lineage (Hollister et al 2012, Yant et al 2013; Bohutínská et al 2021). In these analyses we found genome-wide outliers for several different metrics and this allowed us to identify regions that are very strongly differentiated between diploids and tetraploids, and low diversity in tetraploids (which suggests they were targets of natural selection in the tetraploid lineage; Figure 5). Meiosis genes are strongly over-represented in these lists, fitting with the observation that meiosis is strongly challenged by whole genome duplication. But do the novel alleles of these genes fix the neo-polyploid meiotic problems, and if so, how? A convenient way to study functional differences among alleles is to generate genetic lines that segregate the alternate (ancestral diploid vs. derived tetraploid) alleles in an otherwise tetraploid context. We have recently done this for three of the meiotic genes under selection. In one study, we bred plants that were homozygous for alternate alleles of meiotic axis proteins (ASY1 and ASY3), and compared them using detailed cytological analyses of metaphase I and prophase I (Morgan et al. 2020). From this we learned that the derived alleles of the two axis proteins, one (ASY1) in particular, do affect meiotic traits in a measurable way – and the tetraploid allele was specifically associated with a lower multivalent rate. The interacting partner (ASY3) shows a similar but weaker effect. Thus, we conclude that these two proteins targeted by selection do in fact contribute to the stabilization of tetraploid meiosis. A follow-up study addressed the role of another gene, REC8, which encodes a meiosis-specific subunit of the cohesin complex. In this case, the tetraploid-specific allele is associated with reduced univalent rates (another important adaptation of the evolved polyploids), but we found complex interactions between REC8 and the derived vs ancestral alleles of ASY1 and ASY3, which led us to suggest a model in which REC8 likely evolved first to address the univalent problem, and this then paved a path that allowed the evolution of novel alleles of ASY1 and ASY3, which seem to be deleterious when REC8 is in the ancestral state, but not when it is in the derived state (Morgan et al. 2022).
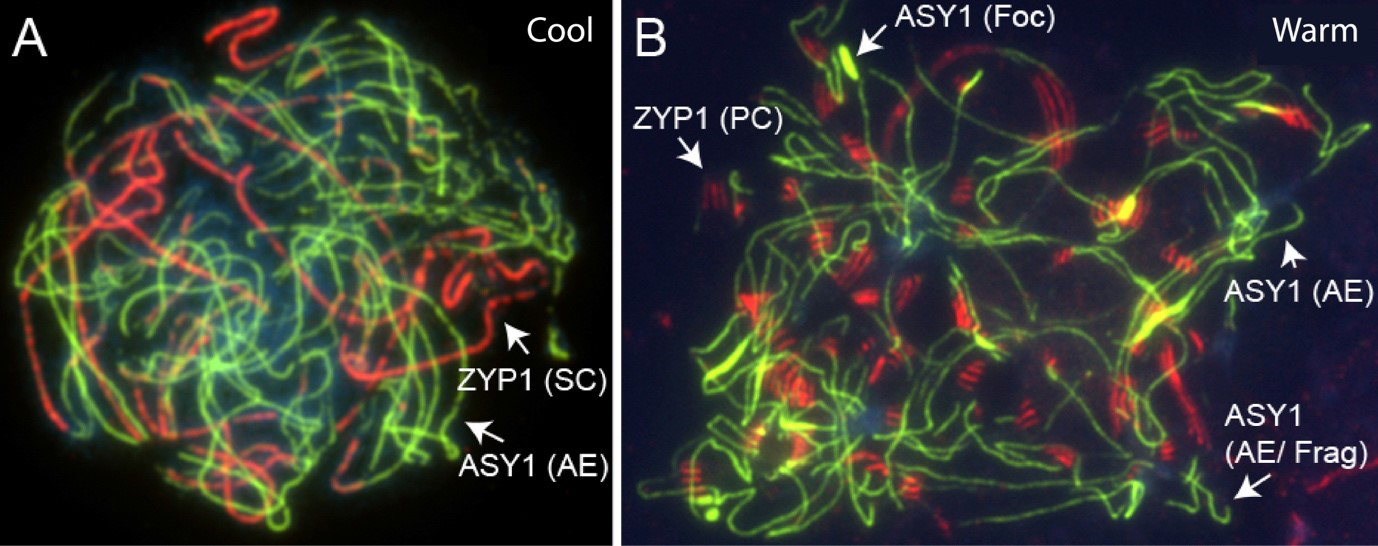
Meiotic adaptation to temperature: An additional study we did examining selection in genomes of diploid A. arenosa lineages, uncovered a second example of selection acting on two meiosis genes (Wright et al. 2015). These genes (the axis component ASY3 and the meiotic cohesin subunit REC8) were also under selection in the tetraploids, but the alleles were entirely distinct from those under selection in tetraploids, showing these were two independent events. Why would selection target meiotic axis and cohesin genes in a diploid? Our current hypothesis is that it has to do with adaptation to temperature, an idea we have been developing for years (Bomblies et al. 2015; Morgan et al. 2017). The hypothesis is based on several facts related to meiosis and the population of diploids where we found selection: (1) meiosis is temperature sensitive, and when plants experience high temperature stress, numerous abnormalities in axis formation and synapsis appear (Figure 6), (2) recombination rates, which can be affected by cohesin, are plastic to temperature (Lloyd et al. 2018), and may reflect an early indication that meiosis is becoming unstable (Morgan et al. 2017), (3) the diploid populations where meiosis is under selection are from the Pannonian Basin in Hungary, which is hotter and drier than where other A. arenosa are found (Wright et al. 2015), (4) we found previously that there might be fertility differences at different temperatures and that the Pannonian populations are more thermotolerant. As part of an SNSF-funded project, we are currently working to understand the adaptation of meiosis to temperature from several angles, but are mostly focused on population variation in recombination and how this relates to thermal stability of meiosis.
Pollen Tubes
We previously showed (see meiosis section) that we could partly ameliorate meiotic defects in A. thaliana neo-polyploids (Gonzalo et al 2023). However, this did not rescue fertility, suggesting that either we did not rescue meiosis enough, or that there is something more. We set out to explore whether it might be the latter, by re-examining our genes under selection. There we found two genes involved in pollen tube tip-growth. Tip-growth is a type of polar cell growth that allows cells to extend hundreds or thousands of times their initial length – it occurs e.g, in neurons, fungal hyphae, and in the pollen tubes of seed plants, where they are essential for fertility because the pollen tubes deliver the sperm to the ovules buried deep in maternal tissues.
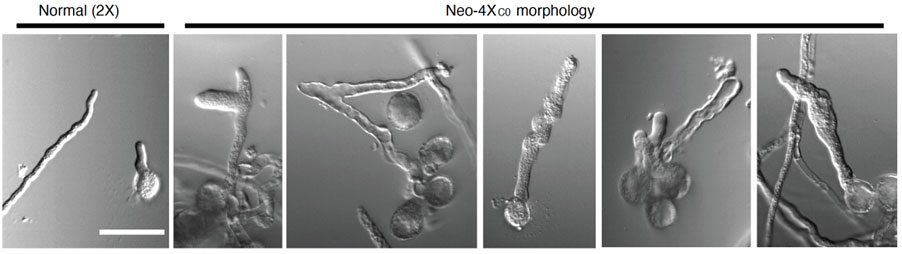
Tip-growth requires tight control of vesicle trafficking, control of cell wall composition and flexibility, and tight control of ion gradients in the growing tip. The genes we found under selection (AGC1.5 and ACA8) are implicated in these processes. We thus sought to test (1) whether neo-polyploids in fact have a problem with pollen tube growth, and (2) whether these genes are involved in stabilizing them. We found evidence for both: Neo-polyploids do have unstable pollen tubes, which, when grown in vitro, commonly burst and/or show morphological abnormalities (Figure 7). In vivo, most fail at the transition point between the dense stigma tissue into the open transmitting tract. The established polyploids do not have these problems. The issues are associated with disruptions in the cytoskeleton, and disruption of the tip-focused Calcium gradient. We then showed, using genetic association, that the tetraploid alleles of AGC1.5 and ACA8 additively help solve these problems and can explain most of the difference between neo-tetraploids and evolved tetraploids. This work was recently published in Science (Westermann et al 2024).
In follow-up work involving collaborations with Thomas Micheals (ETH), Julia Santiago (University of Lausanne), and Ueli Grossniklaus (Univeristy of Zürich), we are using modelling as well as detailed phenotyping to better understand what exactly causes the defects we see in the neo-tetraploids, and how the novel alleles at AGC1.5 and ACA8 help fix the issue. At the same time, we are testing additional candidate genes – including additional ion transporters and other candidates.
Chromatin and the nucleotypic effect
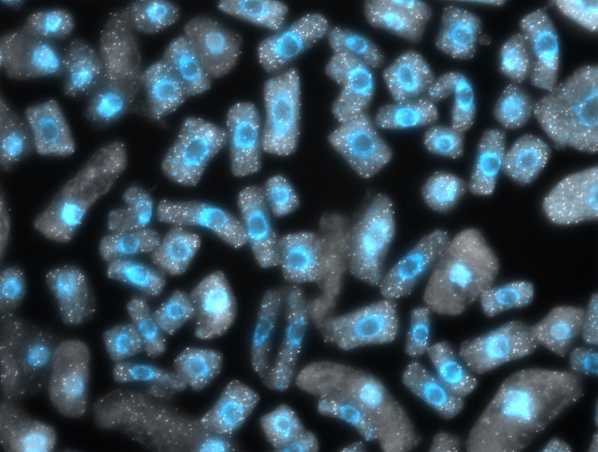
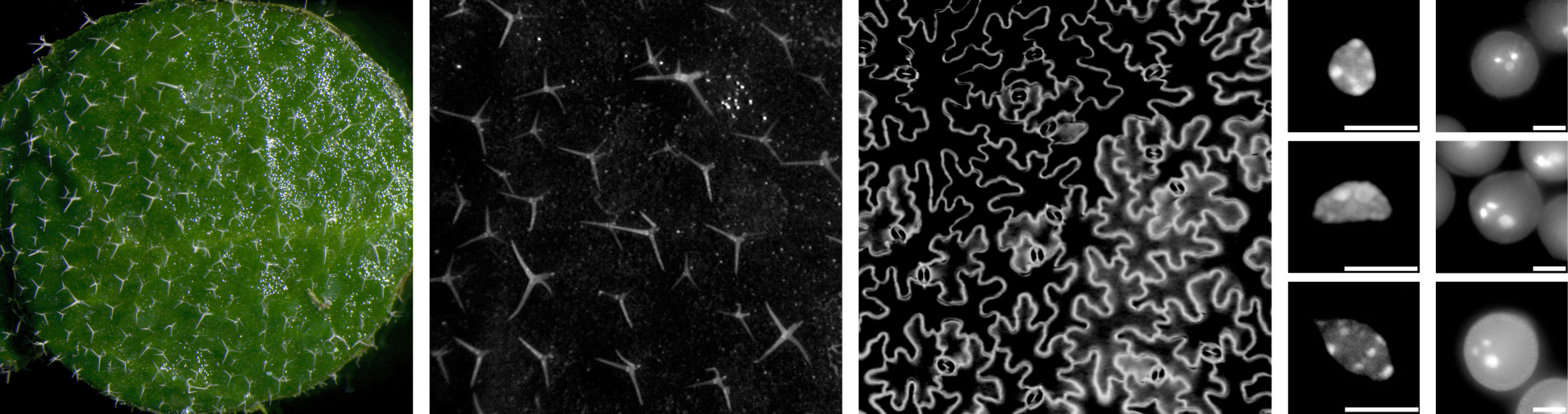
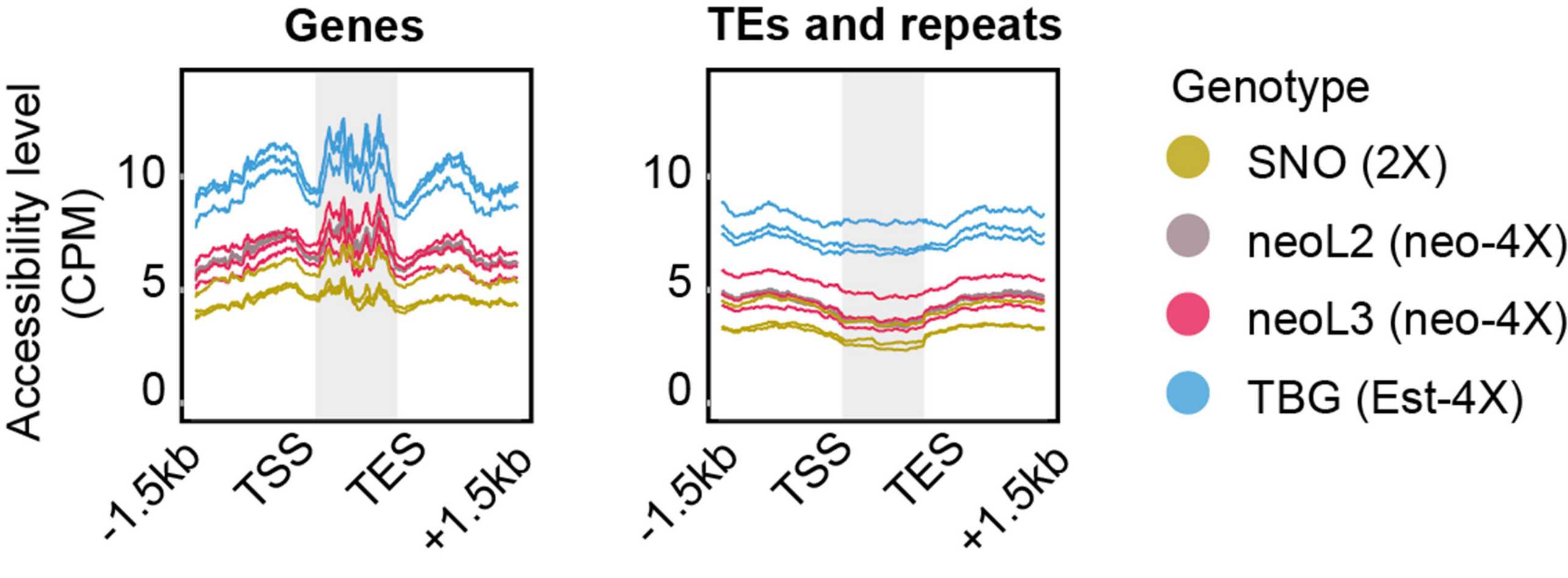
There is an extensive literature describing the so-called “nucleotypic effect”, which, loosely put, refers to the fact that the cell size increase that accompanies whole genome duplication causes scaling mismatches within cells, and challenges associated with dealing with larger volumes. Thus, it is interesting that we, like others working in other species, have observed that cell size, while it increases with whole genome duplication, declines again over evolutionary time. This observation supports the idea that increased cell size really does present a problem. It is clearly not an existential problem, in the sense that neo-polyploids survive, but it does seem to be something that should be undone. Why and how cell size is reduced, and the implications of the changes associated with it, frames our third major research area, which is focused on chromatin organization and how it is affected by, and/or drives, changes in nuclear volume. We are currently using cell biological (Figure 8) and genomic (Figure 9) approaches to better study this process and its knock-on effects for gene regulation in polyploids.
The lab’s publications cited above that have particular relevance to our current research (reverse chronological order):
Westermann, J., Gonzalo, A., Srikant, T., Tan, H. S., Bomblies, K. (2024) Defective pollen tube tip-growth induces neo-polyploid infertility. Science 383, eadh0755 (2024). DOI: 10.1126/science.adh0755 external page ⇒
Gonzalo, A., Parra-Nunez, P., Bachmann, A., Sanchez-Moran, E., Bomblies, K. (2023) Partial cytological diploidization of neo-autotetraploid meiosis by induced crossover rate reduction. Proc Natl Acad Sci USA 120: e2305002120. external page ⇒
Morgan, C., Knight, E., Bomblies, K. (2022) The meiotic cohesin subunit REC8 contributes to multigenic adaptive evolution of autopolyploid meiosis in Arabidopsis arenosa. PLoS Genetics 18: e1010304. external page ⇒
Morgan, C.*, White, M. A.*, Franklin, F. C. H., Zickler, D., Kleckner, N., Bomblies, K. (2021) Evolution of crossover interference enables stable autopolyploidy by ensuring pairwise partner connections in Arabidopsis arenosa. Current Biology 31:R1442-1444 *Contributed equally. external page ⇒
Bohutínská, M., Handrick, V., Yant, L., Schmickl, R., Kolár, F., Bomblies, K.*, Paajanen, P.* (2021) De-novo mutation and rapid protein (co-)evolution during meiotic adaptation in Arabidopsis arenosa. Mol Biol Evol. 38: 1980-1994. *Co-corresponding authors. external page ⇒
Morgan, C.*, Zhang, H.*, Henry, C. E., Franklin, F.C.H., Bomblies, K. (2020) Derived alleles of two axis proteins affect meiotic traits in autotetraploid Arabidopsis arenosa. Proc Natl Acad Sci USA 117: 8980-8988. *Co-first authors external page ⇒
Lloyd, A.*, Morgan, C.*, Franklin, F.C.H., Bomblies, K. (2018) Plasticity of meiotic recombination rates in response to temperature in Arabidopsis. Genetics 208: 1409-1420. *Co-first authors. external page ⇒
Morgan, C.M., Zhang, H., Bomblies, K. (2017) Are the effects of elevated temperature on meiotic recombination and thermotolerance linked via the axis and synaptonemal complex? Invited review. Proc Roy Soc London B. 372: 20160470. external page ⇒
Bomblies, K., Jones, G.H., Franklin, F.C.H., Zickler, D., Kleckner, N. (2016) The challenge of evolving stable polyploidy: could interference play a central role? Invited review. Chromosoma 125: 287-300. external page ⇒
Arnold, B., Kim, S-T., Bomblies, K. (2015) Single geographic origin of a widespread autotetraploid Arabidopsis arenosa lineage followed by interploidy admixture. Mol Biol Evol 32: 1382-1395. external page ⇒
Bomblies, K., Higgins, J.D., Yant, L. (2015) Meiosis evolves: Adaptation to external and internal environments. Invited Tansley Review. New Phytologist 208: 306-323. external page ⇒
Wright, K.M., Arnold, B., Xue, K., Šurinová, M., O’Connell, J., Bomblies, K. (2015) Selection on meiosis genes in diploid and tetraploid Arabidopsis arenosa. Mol Biol Evol 32: 944-955. external page ⇒
Yant, L.*, Hollister, J.*, Wright, K.M., Arnold, B.J., Higgins, J.D., Franklin, F.C.H., Bomblies, K. (2013) Meiotic adaptation to genome duplication in Arabidopsis arenosa. Current Biology 23: 2151-2156. * = *Co-first authors. external page ⇒
Hollister, J., Arnold, B., Svedin, E., Xue, K. Dilkes, B., Bomblies, K. (2012) Genetic adaptation associated with genome-doubling in autotetraploid Arabidopsis arenosa. PLoS Genetics 8: e1003093. external page ⇒
See all publications: ⇒