Chromatin accessibility and gene expression vary between a new and evolved autopolyploid of Arabidopsis arenosa
Thanvi, Adrian and Kirsten from our group just published a Molecular Biology and Evolution paper, describing how epigenomic and transcriptomic changes occur during polyploidy evolution in plants.
They used the lab’s favourite model system Arabidopsis arenosa to examine the immediate consequences of whole-genome duplication (WGD) at the molecular level and how these change over evolution. Performing ATAC-seq (a sequencing approach to assay genome-wide chromatin accessibility) on leaf and petal samples of three cytotypes (diploids, neo-tetraploids and evolved autotetraploids), they identified more than 8000 regions in the genome that change in accessibility levels, which are largely tissue-specific. These regions showed distinct trends across cytotypes, with roughly 70% arising at the immediate onset of WGD. RNA-seq analyses on the same samples (to measure gene expression levels) showed that only a quarter of all genes were differentially expressed across cytotypes. However, ~60% of these genes exhibited large expression changes that reverted to the diploid state over evolution, suggesting that most initial perturbations don’t last. Interestingly, regions that exhibit differential chromatin accessibility show minimal positional overlaps with differentially expressed genes, but are highly enriched for transposable elements. Differentially accessible regions also exhibit higher changes across cytotypes when they occur in proximity to multiple transposable elements. Taken together, these results provide new insights on how epigenomic and transcriptional mechanisms can be dynamic during the evolution of autopolyploidy in plants.
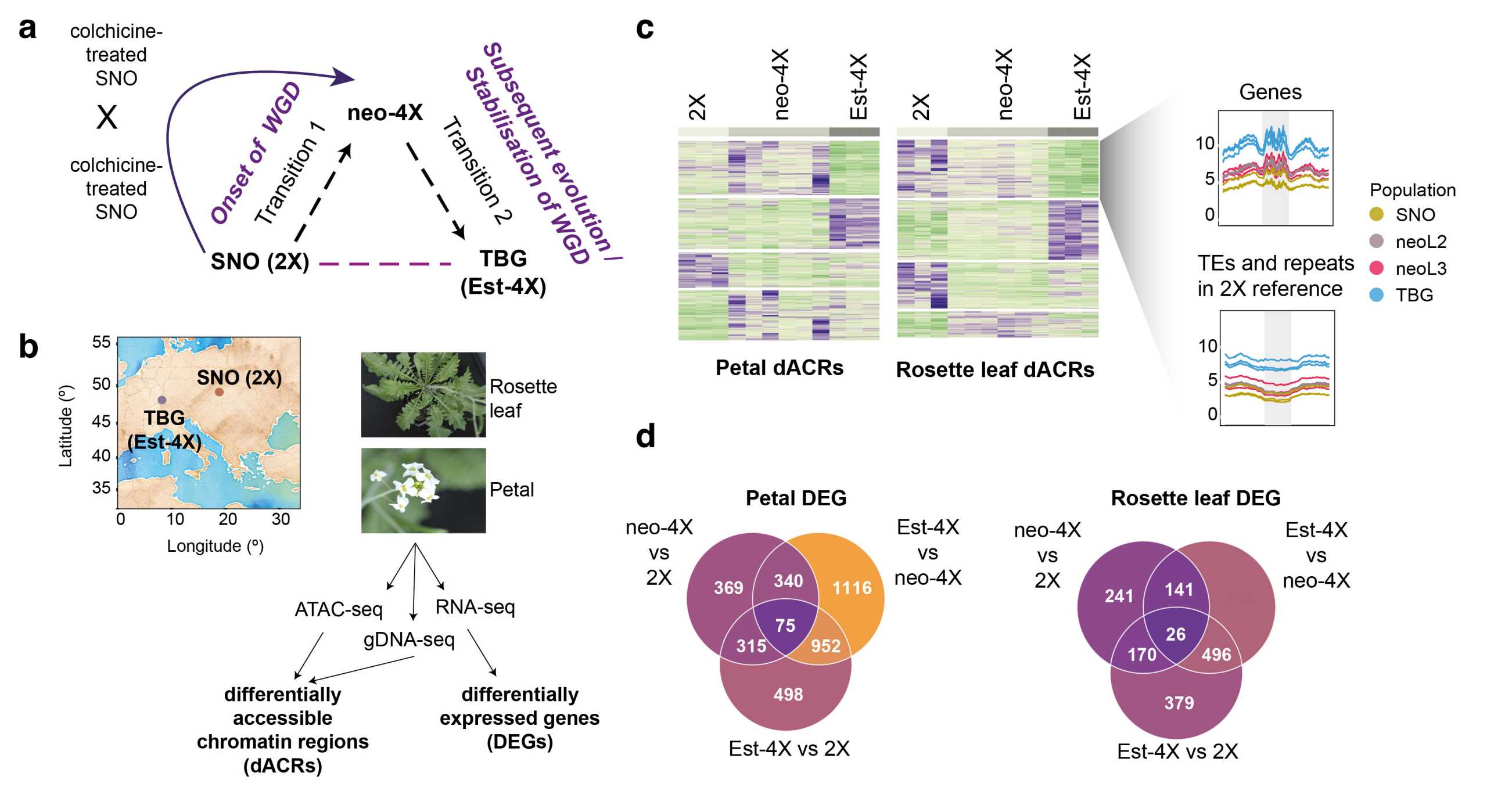
Chromatin accessibility and transcriptome changes during polyploid evolution in A. arenosa. (a) Comparison of three cytotypes for this study to understand the immediate and evolutionary consequences of whole-genome duplication (WGD). Diploids are indicated as 2X, neo-tetraploids as neo-4X, and established or evolved tetraploids as Est-4X. (b) Geographical origins of the diploid and autotetraploid populations used in this study and the plant tissues used to generate sequencing libraries. (c) Changes in chromatin accessibility levels across cytotypes show distinct trends, especially over genes and transposable elements. (d) Numbers of differentially expressed genes (DEGs) identified in various comparisons across cytotypes.
Link to paper external page Molecular Biology and Evolution